 We are pleased to announce our new review paper describing challenges in amplicon and metagenomics sequencing, which has just been published in Briefings in Bioinformatics (https://doi.org/10.1093/bib/bbz155).
We are pleased to announce our new review paper describing challenges in amplicon and metagenomics sequencing, which has just been published in Briefings in Bioinformatics (https://doi.org/10.1093/bib/bbz155).
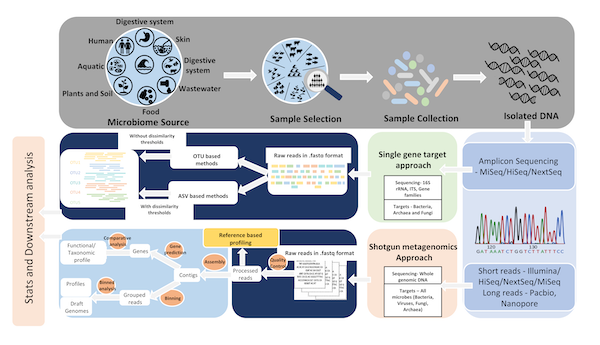
Microbiome research has revolutionized the scientific landscape as a recurrent topic covered in many different disciplines, including animal and plant biology, as well as disease and environmental biology. However, the analyzing the microbiome is highly affected by experimental conditions and computationally intensive downstream analyses. The aim of this work is to review the technical challenges in microbiome research and to provide a best practice workflow for an optimal microbiome analysis. The paper comprehensively reviews commonly used experimental and computational workflows of 16S rRNA sequencing, shotgun and long read metagenomics. The best practice workflows for 16S rRNA and metagenomic sequencing data are released as bash scripts at: https://github.com/grimmlab/MicrobiomeBestPracticeReview
More details can be found in our publication:
Bharti and D.G. Grimm, “Current challenges and best-practice protocols for microbiome analysis.” Briefings in Bioinformatics, 2019
https://doi.org/10.1093/bib/bbz155
