Abstract
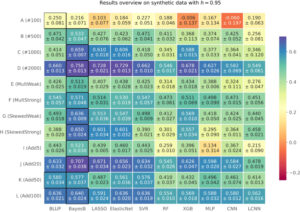
Genomic selection is an integral tool for breeders to accurately select plants directly from genotype data leading to faster and more resource-efficient breeding programs. Several prediction methods have been established in the last few years. These range from classical linear mixed models to complex non-linear machine learning approaches, such as Support Vector Regression, and modern deep learning-based architectures. Many of these methods have been extensively evaluated on different crop species with varying outcomes. In this work, our aim is to systematically compare 12 different phenotype prediction models, including basic genomic selection methods to more advanced deep learning-based techniques. More importantly, we assess the performance of these models on simulated phenotype data as well as on real-world data from Arabidopsis thaliana and two breeding datasets from soy and corn. The synthetic phenotypic data allow us to analyze all prediction models and especially the selected markers under controlled and predefined settings. We show that Bayes B and linear regression models with sparsity constraints perform best under different simulation settings with respect to explained variance. Further, we can confirm results from other studies that there is no superiority of more complex neural network-based architectures for phenotype prediction compared to well-established methods. However, on real-world data, for which several prediction models yield comparable results with slight advantages for Elastic Net, this picture is less clear, suggesting that there is a lot of room for future research.
Original Publication (Open Access)
John Maura, Haselbeck Florian, Dass Rupashree, Malisi Christoph, Ricca Patrizia, Dreischer Christian, Schultheiss Sebastian J., Grimm Dominik G. (2022).
A comparison of classical and machine learning-based phenotype prediction methods on simulated data and three plant species. Frontiers in Plant Science, 2022. https://doi.org/10.3389/fpls.2022.932512
