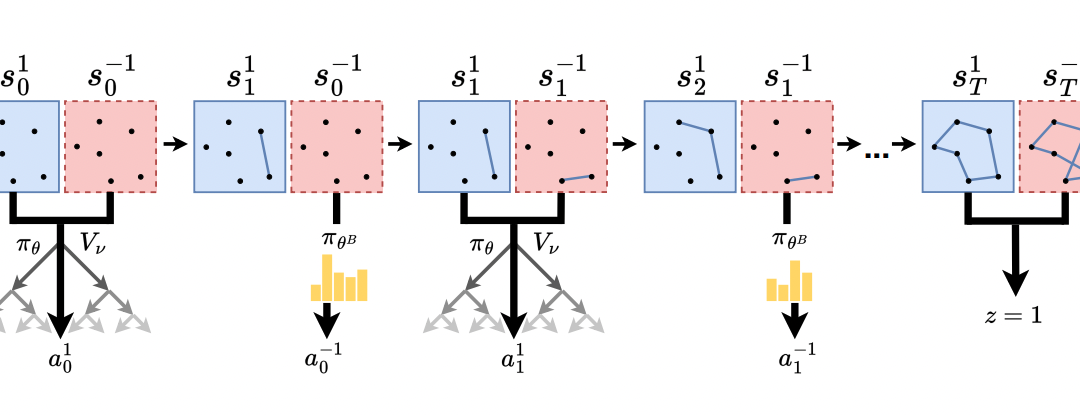
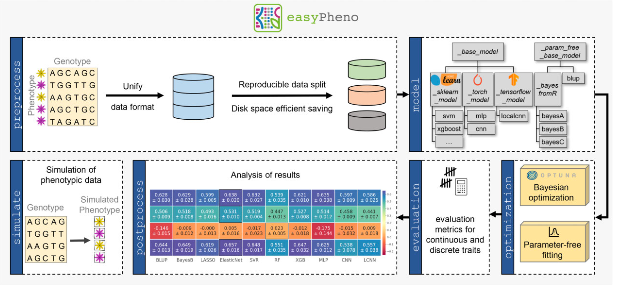
Machine Learning Research School in Bangkok: Poster Award für Josef Eiglsperger
Award for Josef Eiglsperger: The doctoral student at the Professorship of Bioinformatics (Prof. Dominik Grimm) at Weihenstephan-Triesdorf University of Applied Sciences (HSWT) at TUM Campus Straubing (TUMCS) received the “Outstanding Poster Award” for his poster and presentation at the Machine Learning Research School (MLRS) in Bangkok. The Research Summer School ran from 02.08. to 09.08.2023 and took place at the Digital Economy Promotion Agency.