Abstract
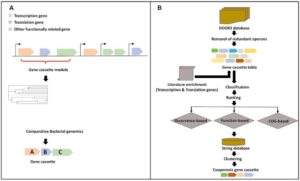
Transcriptional-translational coupling is accepted to be a fundamental mechanism of gene expression in prokaryotes and therefore has been analyzed in detail. However, the underlying genomic architecture of the expression machinery has not been well investigated so far. In this study, we established a bioinformatics pipeline to systematically investigated >1800 bacterial genomes for the abundance of transcriptional and translational associated genes clustered in distinct gene cassettes. We identified three highly frequent cassettes containing transcriptional and translational genes, i.e. rplk-nusG (gene cassette 1; in 553 genomes), rpoA-rplQ-rpsD-rpsK-rpsM (gene cassette 2; in 656 genomes) and nusA-infB (gene cassette 3; in 877 genomes). Interestingly, each of the three cassettes harbors a gene (nusG, rpsD and nusA) encoding a protein which links transcription and translation in bacteria. The analyses suggest an enrichment of these cassettes in pathogenic bacterial phyla with >70% for cassette 3 (i.e. Neisseria, Salmonella and Escherichia) and >50% for cassette 1 (i.e. Treponema, Prevotella, Leptospira and Fusobacterium) and cassette 2 (i.e. Helicobacter, Campylobacter, Treponema and Prevotella). These insights form the basis to analyze the transcriptional regulatory mechanisms orchestrating transcriptional–translational coupling and might open novel avenues for future biotechnological approaches.
Original Publication (Open Access)
Richa Bharti, Daniel Siebert, Bastian Blombach, Dominik G Grimm
Systematic analysis of the underlying genomic architecture for transcriptional–translational coupling in prokaryotes, NAR Genomics and Bioinformatics, Volume 4, Issue 3, September 2022, lqac074, https://doi.org/10.1093/nargab/lqac074
