
Sofia joins the Team as Research Assistant
Sofia joins the team as research assistant. She will work on novel machine learning methods for synthetic protein design and the in silico evaluation of generated artificial sequences.

Sofia joins the team as research assistant. She will work on novel machine learning methods for synthetic protein design and the in silico evaluation of generated artificial sequences.

We are delighted to have received funding from the European Innovation Council (EIC) as part of the EIC Transition programme to work with colleagues at the Technical University of Munich (TUM) to develop a new technology that will make it quicker, easier and more accurate to apply fertiliser in the field. Using a combination of biosensor test strips and satellite-based remote sensing data, the Technical University of Munich (TUM) is developing a method to determine the nutritional status of cereals and the perfect amount of fertiliser to apply. The automatic provision of digital analysis data to the tractor terminal should save time and prevent over-fertilisation in the future. The Straubing Campus for Sustainability plays a leading role within the TUM in the development of environmentally friendly technologies. The GrimmLab will be responsible for the development of the machine learning related parts for fertilizer recommendations.
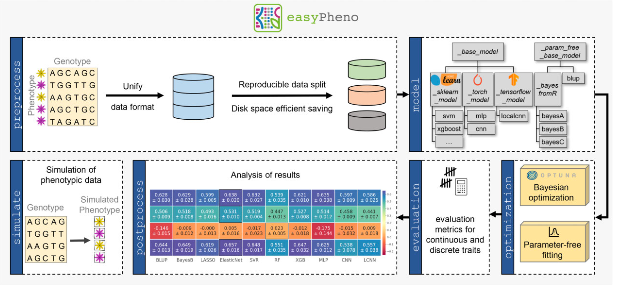
New paper in Bioinformatics Advances: “easyPheno: An easy-to-use and easy-to-extend Python framework for phenotype prediction using Bayesian optimization”. Predicting complex traits from genotypic information is a major challenge in various biological domains. With easyPheno, we present a comprehensive Python framework enabling the rigorous training, comparison and analysis of phenotype predictions for a variety of different models, ranging from common genomic selection approaches over classical machine learning and modern deep learning-based techniques. Our framework is easy-to-use, also for non-programming-experts, and includes an automatic hyperparameter search using state-of-the-art Bayesian optimization. Moreover, easyPheno provides various benefits for bioinformaticians developing new prediction models. easyPheno enables to quickly integrate novel models and functionalities in a reliable framework and to benchmark against various integrated prediction models in a comparable setup. In addition, the framework allows the assessment of newly developed prediction models under pre-defined settings using simulated data. We provide a detailed documentation with various hands-on tutorials and videos explaining the usage of easyPheno to novice users.
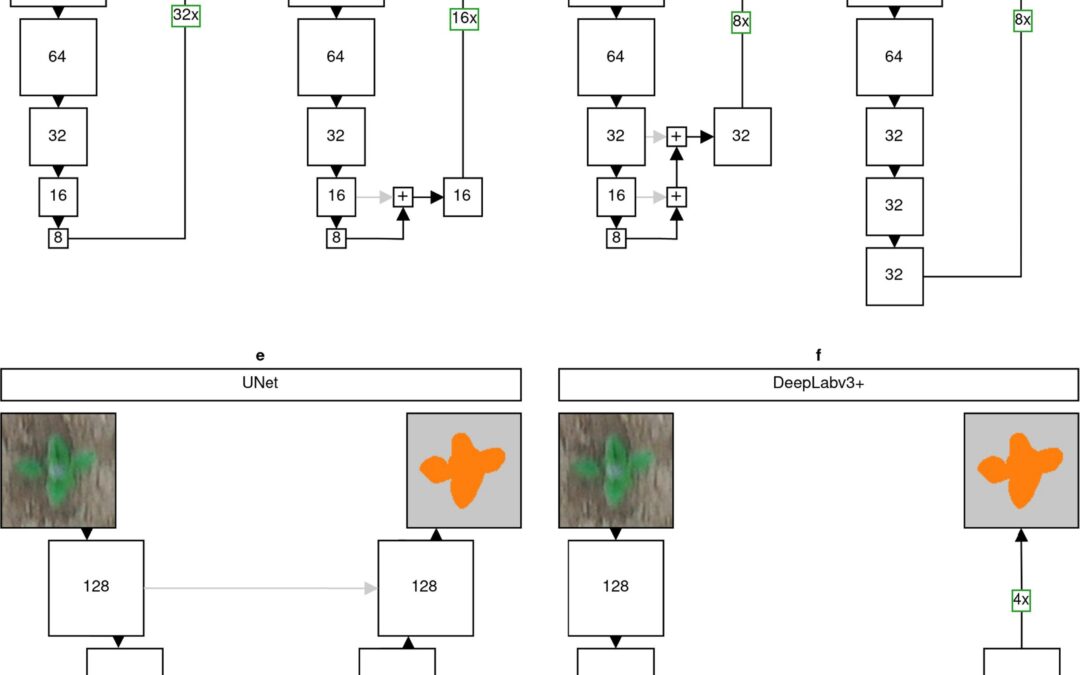
We are happy to receive funding from the Bavarian State Ministry of Food, Agriculture and Forestry to continue or research about developing novel machine learning techniques for weed identification in drone imagery to enable automatic weed removal due to autonomous robots on agricultural fields.

In this episode Dominik gives us insights into CropML, a BMBF funded project. The project evaluates new machine learning techniques for more accurate plant breeding by integrating heterogeneous external factors. Different phenotype prediction models, including basic genomic selection methods to more advanced deep learning-based techniques have been compared. Learn why advanced models are the future and where the challenges are.